Organitzat per l'IRB Barcelona en col·laboració amb la Fundació Catalunya – La Pedrera.
Els tutors d'aquesta edició són: Sanja Zivanovic, Ricardo Viais, Nina Schweizer, Mark McCully, Núria Gutiérrez, Jordi Badia, Marion Salzer, Jorge García, Susanna Barros, Daniela Kalafatovic, Lada Murcia i Celia Santos.
Presentation

Participants del curs "Crazy About Biomedicine", edició 2017.
Objectius
Bojos per la Biomedicina és un curs dirigit als estudiants del primer any de batxillerat que desitgin explorar alguns dels descobriments fascinants que s'estan fent actualment en les ciències de la vida. A través d'aquest curs, els estudiants tindran l'oportunitat d'aprofundir el seu coneixement de la teoria i tècniques científiques en el camp de la biomedicina. Treballaran juntament amb investigadors joves per experimentar com es fa ciència en un institut de recerca internacional, guanyar una mica d'experiència pràctica en les últimes metodologies d'avantguarda i posicionar-se per a una possible carrera professional en les ciències de la vida.
Descripció del curs
Taller d'un any de durada sobre les ciències de la vida per a estudiants de batxillerat. Organitzat per IRB Barcelona dins el Programa "Bojos per la Ciència" de la Fundació Catalunya – La Pedrera.
Aquest curs combina sessions teòriques i activitats experimentals pràctiques, que es duran a terme durant 18 dissabtes de l'any. El curs tractarà 12 temes científics actuals, que van des de la biologia cel·lular i molecular fins a la biologia estructural i computacional i la química, presentats per estudiants de doctorat i postdocs de l'IRB Barcelona. En el primer «semestre» (gener-juny 2017), els 3 primers dissabtes es dedicaran a aquestes sessions teòriques generals per a tots els participants. Durant els 6 dissabtes següents, es formaran grups petits que entraran als laboratoris per a les sessions pràctiques. A continuació, es repetirà aquest programa amb 6 temes de recerca nous per al segon semestre (juny-desembre). Els estudiants participants s'hauran de comprometre a assistir al curs durant tot l'any.
 Idioma del curs
Idioma del curs
Totes les xerrades i sessions pràctiques es faran en anglès.
Dates i horaris
El curs es farà de gener a desembre de 2017, 10.00h-14.00h.
1r SEMESTRE
- Div. 13 gener 2017: Inauguració al Mon Sant Benet- Sant Fruitós del Bages
- Diss. 14 gener: Xerrada 1
- Diss. 21 gener: Xerrada 2
- Diss. 4 febrer: Xerrada 3
- Diss. 18 febrer: Taller 1
- Diss. 4 març: Taller 2
- Diss. 18 març: Taller 3
- Diss. 1 abril: Taller 4
- Diss. 22 abril: Taller 5
- Diss. 29 abril: Taller 6
2n SEMESTRE
- Diss. 20 maig: Xerrada 1
- Diss. 3 juny: Xerrada 2
- Diss. 17 juny: Xerrada 3
- Diss. 16 setembre: Taller 1
- Diss. 30 setembre: Taller 2
- Diss. 21 octubre: Taller 3
- Diss. 28 octubre: Taller 4
- Diss. 11 novembre: Taller 5
- Diss. 25 novembre: Taller 6
Preu del curs
Lloc on es realitzarà el curs
Institut de Recerca Biomèdica (IRB Barcelona)
C/ Baldiri Reixac, 10
08028 Barcelona
Qui pot sol·licitar una plaça
Aquest curs està dirigit als estudiants de primer any de batxillerat que tinguin un interès i talent especials en els camps relacionats amb les ciències de la vida (principalment biologia i química).
Els estudiants poden sol·licitar la plaça a un màxim de 2 dels programes de la série "Bojos per la Ciència" i participar només en un.
Com sol·licitar una plaça

Els estudiants interessats hauran d'emplenar el formulari web de sol·licitud i incloure una carta de motivació. També es demanarà una carta de recomanació directament de dos dels seus professors que coneguin bé l'alumne/a. En el cas de que l'estudiant hagi canviat de centre aquest curs, suggerim que sol·licitin les cartes als antics professors. La data límit d'inscripció és el 23 d'octubre 2016.
El curs està obert a un total de 24 estudiants. Se seleccionaran els candidats en funció del seu expedient acadèmic, de les recomanacions dels seus professors i de la seva motivació per participar-hi. Es convidarà els candidats preseleccionats a fer entrevistes amb els organitzadors científics al novembre, després de les quals es farà la selecció final. La primera setmana de desembre es comunicarà el resultat als estudiants. Es demanarà als estudiants seleccionats per participar-hi i als seus pares/tutors legals que signin una carta de compromís d'assistir a totes les sessions.
Col·laboradors
Facebook: @catfundacio
Twiter: @catfundacio
Instagram: #bojosperlaciencia @catfundacio
Facebook: @youthscience.barcelona
Twiter: @youthsciencebcn
Instagram: @youthscience.barcelona



Per a qualsevol dubte, siusplau contacteu amb: irb_outreach@irbbarcelona.org
Dates importants
 23 d'octubre 2016: Data límit d'inscripció
23 d'octubre 2016: Data límit d'inscripció- Setmana del 7 de novembre 2016: Contacte amb els candidats preseleccionats
- 14-25 de novembre 2016: Entrevistes
- Setmana del 28 de novembre 2016: Contacte amb els candidats seleccionats
- 1-5 de desembre 2016: Publicació del llistat dels estudiants acceptats
- Div. 13 de gener 2017: Inauguració del curs al Món Sant Benet –Sant Fruitós del Bages
- Diss. 14 de gener 2017: Inici del curs
- Dill. 27 de novembre 2017: Cerimònia de clausura
Programme
SEMESTER 1
1. Rational drug design
Sanja Zivanovic (Molecular modelling and bioinformatics)

The typical drug discovery and development cycle, from concept to market, takes approximately 14 years and can cost up to 1 billion euros. This process could be vastly improved by combining scientific knowledge and powerful computational methods. Such an approach aims to identify the damaged protein that causes illness and also find hits and possible inhibitors, thus reducing the labour costs and the time-consuming process of drug discovery.
Students will be introduced to the fundamentals of molecular modelling and will have the opportunity to practise drawing 2D and 3D structures. They will learn how to manipulate atoms and molecules, set up correct charge, and analyse optical activity. We will go through various molecular visualization software packages, namely Avogadro, VMD, and Pymol, and perform graphical representations of molecules. To be able to use the above-mentioned software, students will be provided with the necessary computational tools such as a command line in Linux. The skills acquired in this tutorial will help them to carry out more drug discovery calculations in the second semester.
2. Stepping into the world of the cell skeleton
Ricardo Viais and Nina Schweizer (Microtubule organization)

The cytoskeleton is a cellular compartment present in all domains of life. As its name suggests, it is the “skeleton” of the cell, ensuring its shape and migration. In eukaryotic cells, one of the components of the cytoskeleton is the microtubule—a tiny tubular structure that forms a network throughout the cytoplasm. Microtubules can be considered LEGO, where the basic individual bricks are dimers of a protein called tubulin. The microtubule network is extremely dynamic, and the addition or removal of tubulin “pieces” from microtubules leads to their growth or shrinkage. In fact, microtubules are much more than just a “skeleton of the cell”. They are also involved in chromosome segregation during mitosis and meiosis and they serve as highways for the transport of organelles and key molecules inside the cell. Microtubules are
 one of the targets of cancer treatment and have also been shown to be involved in neurodegenerative diseases (like Parkinson’s and Alzheimer’s).
one of the targets of cancer treatment and have also been shown to be involved in neurodegenerative diseases (like Parkinson’s and Alzheimer’s).
In this practical course we will learn how microtubules are formed, how they grow, and what they look like in different cell types and distinct cell cycle stages. For this purpose, we will use cultured cell lines, mouse hippocampal neurons, and fluorescence light microscopy techniques.
3. Gateway to the brain: Creating peptides to cross the blood brain barrier and deliver therapeutics
Mark McCully (Design, synthesis and structure of peptides and proteins)

The human body has numerous barriers that serve to ensure a stable environment for our cells and to protect us from disease. Our skin, the mucous lining in respiratory system, and tears from our eyes are all examples of barriers designed to safeguard our wellbeing.
The brain is an extremely complicated organ and as such it too has a highly complex barrier to shield it against viruses, bacteria and chemicals. Called the Blood-Brain Barrier (BBB), this system is of the utmost importance to keep our brains healthy. However, the BBB is not totally impenetrable, and some external agents are able to cross it and invade the brain. This is the point at which problems truly start for doctors and their patients. The BBB is so complex and selective that sending traditional drugs to target the disease does not work. Therapeutic agents simply do not have the capacity to cross the BBB, thus resulting in high mortality rates for brain diseases.
Our lab is focused on developing peptides (short sequences of amino acids) that can cross this barrier and transport a drug of interest to the site of the infection. During this course we will synthesise peptides, attach a “therapeutic” cargo and assess the capacity of these constructs to cross the BBB.
4. Study of therapeutic drugs that trigger apoptosis in cancer cells
Núria Gutiérrez (Signalling and Cell Cycle Laboratory)

Apoptosis is a form of programmed cell death that is required for organisms to grow and develop properly and to maintain body tissues. This process is initiated when a cell is damaged or infected. DNA damage or oncogene expression, among other stimuli, can induce cell cycle arrest through the activation of special proteins involved in cell cycle checkpoints. When arrested, DNA damage can be repaired and the cell cycle can re-start. If damage persists, cells undergo apoptosis.
A hallmark of human cancer is the intrinsic or acquired resistance to cell death. Cancer cells are able to bypass apoptosis by altering the mechanisms that detect damage or abnormalities. This means that proper signalling does not occur. As a result, unrepaired damage accumulates, giving rise to mutations that allow cells to become malignant. This knowledge has been exploited for the purpose of therapy design. Several anti-cancer drugs impair the DNA damage response, thereby leading to the accumulation of cytotoxic damage, which eventually gives rise to apoptosis. However, different kinds of cancer show distinct behaviours, and the response to these chemotherapeutic drugs varies in function of the cell type.
In this practical course we will treat various cancer cell lines with several chemotherapeutic agents used in clinical practice and test their responses. To detect cell death, we will use one of the most famous techniques in molecular biology, namely Western blot. In parallel, we will learn about other techniques used in this field such as immunofluorescence and immunohistochemistry. These techniques will allow us to appreciate the differences between distinct cancer cells and why therapy design is such a complex process.
5. Targeting the Tumour Microenvironment in colorectal cancer metastasis
Jordi Badia (Colorectal cancer laboratory)

Colorectal Cancer (CRC) is the most common cancer and the second leading cause of cancer-related deaths in Spain, according to the AECC (Asociación Española Contra el Cáncer). CRC produces cancers in the large intestine through a multi-step process that, like many other types of cancer, starts with the acquisition of mutations in key genes. These mutations then go on to make cells proliferate without control. CRC can be easily treated if detected early. However, we still need to find effective treatments for the most advanced stages of cancer, particularly for metastasis—the final and deadliest step of cancer, which in the case of CRC occurs mainly in the liver.
The great potential of cancer cells resides not only in their capacity for mass proliferation but also in their ability to trick the healthy cells surrounding them. Fibroblasts, cells of the immune system, and blood vessels form what we call the tumour microenvironment (TME), which protects cancer cells from being removed and facilitates their invasion of other healthy organs. Our laboratory has recently shown that the TME is crucial for the aggressiveness of CRC and formation of metastasis in the liver. We are now channelling efforts into studying the potential of the TME as a way to block metastasis and into identifying novel treatments.
In this practical course we will provide an overview of several in vivo models and histological tools. Using these tools, we will discover and target those cells that form the TME of liver metastasis, with the aim to learn how to exploit them and stop the CRC metastasis.
6. When cells go wild: Cancer stem cells and their microenvironment
Marion Salzer (Stem cells and cancer)

Throughout life our skin regenerates in order to maintain a functional barrier between our body and the external environment. To do so, epidermal stem cells continuously divide, and their offspring differentiate into functional skin barrier cells, which eventually die and are shed. The processes of stem cell division, differentiation, and death are tightly regulated. Extensive cell division may lead to the formation of a tumour, while extensive cell death compromises the barrier function of skin.
The skin contains a variety of cell types, such as dermal fibroblasts, endothelial cells, and immune cells, which control the behaviour of stem cells by secreting growth-promoting or growth-inhibiting signals. These cells form the so-called “stem cell niche”. Occasionally, a stem cell acquires a mutation that allows it to divide in an uncontrolled manner. If this mutation goes undetected by the microenvironment, it will lead to the development of a tumour. The same cell types that are found in healthy skin are also found in skin tumours, but their behaviour differs. Cancer stem cells hyperproliferate, and the cells in the surrounding niche permit or even enhance this process.
During our practical session we will use cell cultures of primary tumour cells isolated from mice to study the interaction between tumour cells and tumour-associated fibroblasts. We will deplete a gene that is highly expressed in tumour-associated fibroblasts compared to normal fibroblasts and examine its effect on tumour cell behaviour.
SEMESTER 2
1. Regulation of gene expression or why we don't understand our genome (yet)
Manuel Cañete (Translational control of cell cycle and differentiation)

In the year 2000, the Human Genome Project was completed at a cost of around 3 billion dollars and ten years of intense work. The main conclusion was that over 98% of our genetic material was junk DNA. Far from resolving our questions, by reading our genome it became apparent that we didn't understand it. Since then, mounting evidence has made us realise that understanding our genome means understanding how its expression is regulated.
We are now starting to gain some insight into how cells depend on extremely complex regulatory networks to switch genes on and off through a myriad of novel players such as microRNAs, long non-coding RNAs, and RNA-binding proteins. Unravelling the mechanisms underlying the regulation of gene expression will be vital to understand how an erythrocyte and a neuron differ while sharing the exact same genes or why in identical twins one falls ill with a disease like cancer while the other remains completely healthy.
In this course we will use in vitro cell culture systems and look at the current methods that researchers apply to study the regulation of gene expression.
2. Stepping into the world of proteins
Jorge García (Structural biology of protein & nucleic acid complexes and molecular machines)

Proteins form the very basis of life. They regulate a variety of activities in all known organisms, from replication of the genetic code to oxygen transport, and are generally responsible for regulating cellular machinery and determining the phenotype of an organism. Proteins accomplish their tasks in the body by three-dimensional (3D) interactions between various substrates. The functional properties of proteins depend upon their 3D arrangement. The 3D structures arise because particular sequences of amino acids in a polypeptide chain fold to generate from linear chains to compact domains with specific organizations. The folded domains can serve as modules for larger assemblies or provide specific catalytic or binding sites.
Various technologies, such as X-ray crystallography, are used to disclose protein structures. This particular technique calls for the use of crystals, a material whose constituents (in this case proteins) are arranged in an ordered pattern that extend in all three spatial dimensions. Crystals subjected to this technique, scatter the x-rays, and the information collected can help to infer the protein structure.
In this course, we will use the bacteria Escherichia coli to produce a protein of interest. We will use this same protein to make crystals and observe how a real protein crystal looks like!
3. Understanding the relevance of mitochondrial dynamics in metabolic disease
Susana Barros (Complex metabolic diseases and mitochondria)

Age-related metabolic diseases have increased to epidemic proportions over the past century in all industrialized countries. The prevalence of diabetes mellitus and glucose intolerance increases significantly with age. Mitochondrial dysfunction has been reported in complex age-related diseases, as well as in aging itself.
Mitochondria are essential organelles for converting nutrients into usable and storable energy in the form of ATP through different processes such as oxidative phosphorylation, Krebs cycle and fatty acid oxidation. For that mitochondria displays an ability to modulate their architecture through a dynamic process of fusion and fission in order to adapt to the energetic needs of the cell. It had been described that defects in mitochondrial dynamics leads to mitochondrial function impairments contributing to a variety of human diseases.
Preliminary studies in our group showed that the in vivo hepatic ablation of Mfn2, responsible for mitochondrial fusion, led to numerous metabolic abnormalities.
In this practical session students will be introduced to techniques of gene silencing in vitro as a tool to mimic the in vivo model and its relevance in research. Students will also have access to histological studies from different in vivo models and infer differences between normal and knockout mice.
4. Identifying new membrane receptors using combinatorial chemistry
Daniela Kalafatovic (Design, synthesis and structure of peptides and proteins)

Cancer continues to be one of the main causes of death worldwide. Current therapeutic approaches have limitations in that they are not sufficiently specific, effective or localized not are they efficient against metastasis. Metastasis accounts for 90% of all cancer deaths in patients with solid tumours. Preventing metastasis in high risk patients is still the main challenge in cancer medicine.
Recently, specific genomic mutations have been associated with bone metastasis in breast cancer patients. By exploiting this finding, new diagnostic tools for identifying patients at high risk can be developed using a combinatorial chemistry approach. The objective is to identify new membrane receptors associated with specific genomic mutations and clarify the differences between cells with or without metastatic potential. For this purpose, we use peptides bound to resin beads and screen them in various cancer cell lines.
In this course we will address how to build small peptide libraries and how to characterize them. In order to find new binders for cell surface receptors, we will then incubate cancer cells with the peptides and study their interactions.
5. Study of tumorigenesis using flies
Lada Murcia and Celia Santos (Development and Growth Control Laboratory)


Cancer is a multi-hit process that involves mutations in oncogenes and tumour suppressors. It is characterized by the uncontrolled proliferation of malignant cells and invasion to adjacent healthy tissue. The most common type of human tumour, carcinoma, is of epithelial origin. Aggressive cancer cells activate a programme named EMT (Epithelial to Mesenchymal Transition), which causes them to undergo morphogenetic alterations, finally leading to acquisition of invasive motility. Over the last decade, the fruit fly Drosophila melanogaster has become a key model system for cancer research. This model has the advantage that the signalling pathways are conserved between flies and humans, genome redundancy is reduced compared to humans, and a wide range of powerful genetic tools are available.
In this course, we will give an overview of Drosophila genetics and present how we use this model system to characterise aspects of tumorigenesis, such as cell proliferation and cell invasion, at the molecular and cellular levels. In particular, we will focus on the “wing imaginal disc” —simple mono-layered epithelia that grow about 1000-fold in mass and cell number during larval development—. In this regard, we will perform in vivo dissections of this tissue and immunohistochemistry of the samples to analyse the signalling pathways involved in some aspects of tumorigenesis.
6. Study of tumorigenesis using flies (This course is a continuation from the previous)
Lara Barrio (Development and Growth Control Laboratory)

Cancer is a complex disease driven by the accumulation of mutations and epigenetic changes that alter intrinsic cellular functions and cell-cell interactions in tissues. The conservation of signalling pathways and cellular regulatory systems between humans and flies has made Drosophila melanogaster a useful model in which to explore different types of cancer. Indeed, many human oncogenes and tumour-suppressor genes were originally discovered in Drosophila.
In this second part of the course, we will continue to use the "imaginal discs" to study some aspects underlying tumour initiation and progression. We will analyse the previous dissected samples by confocal microscopy, process the images with specific software and finally do a statistical analysis.
Venue
El programa BOJOS PER LA BIOMEDICINA es durà a terme a les instal·lacions de l'IRB Barcelona.
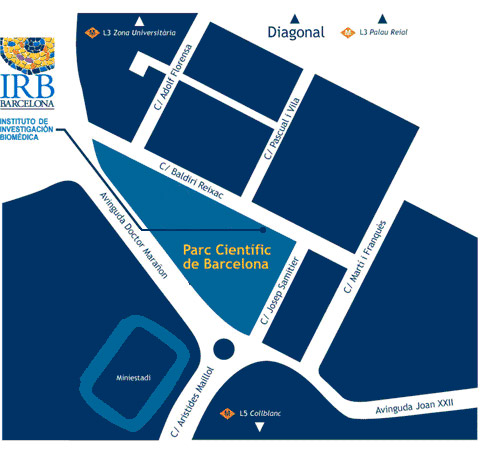
Institut de Recerca Biomèdica (IRB Barcelona)
Parc Cientific de Barcelona
C/ Baldiri Reixac, 10
08028 Barcelona
(Campus de la Diagonal, Universitat de Barcelona)

