Organized by IRB Barcelona in collaboration with the Fundació Catalunya La Pedrera.
Presentation

Objectives
Crazy About Biomedicine is a course directed at students in their first year of baccalaureate who wish to explore some of the exciting discoveries being made in the life sciences. Through this course, students will have a chance to deepen their knowledge of scientific theory and techniques in the field of biomedicine. They will work alongside our young researchers to get a taste for what doing science in a top international research institute is like, gain some hands-on experience in the latest cutting-edge methodologies, and position themselves for a potential career in the life sciences.
Course Description
A year-long workshop in the life sciences for high-school students organized by IRB Barcelona within the series "Crazy About Science" of the Fundació Catalunya La Pedrera.
This course includes a combination of theoretical lectures and practical hands-on experimental activities, which take place on 13 Saturdays throughout the year. Presented by IRB Barcelona PhD students and postdocs, the course will cover 10 hot scientific topics, ranging from cell and molecular biology to structural and computational biology, and chemistry. The first 3 Saturdays will be dedicated o general theoretical sessions for all participants. Over the next 10 Saturdays, groups of 5 students will enter the laboratories for practical sessions. Participants must commit themselves to attend the whole course.
Students will receive a Certificate of Participation upon successful completion of the course at a special ceremony to which parents and teachers will be invited. Moreover, we offer the possibility of performing the practical part of the Treball de Recerca of Batxillerat in our laboratories under the supervision of one of our researchers.
Course language
All lectures and practical sessions will be conducted in English.

Course dates and times
The course will run from January to June 2025, Saturdays 10.00-14.00h.
- Sat. 11 January: Official course inauguration and Talks 1-2-3
- Sat. 25 January: Talk 4-5-6
- Sat. 8 February: Talk 7-8-9-10
- Sat. 15 February: Practical session 1
- Sat. 22 February: Practical session 2
- Sat. 8 March: Practical session 3
- Sat. 22 March: Practical session 4
- Sat. 29 March: Practical session 5
- Sat. 5 April: Practical session 6
- Sat. 26 April: Practical session 7
- Sat. 10 May: Practical session 8
- Sat. 24 May: Practical session 9
- Sat. 7 June: Practical session 10
Course fees
There is a fee of 425 euros payable directly to the Fundació Catalunya La Pedrera. Take a look at the scholarships available at their webpage.
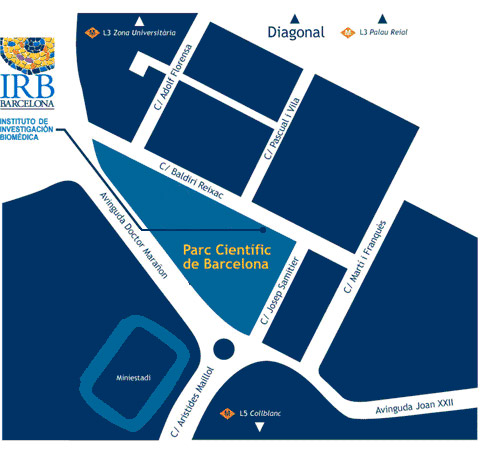
Course location
Institute for Research in Biomedicine (IRB Barcelona)
C/ Baldiri Reixac, 10
08028 Barcelona
Who can apply
This course is directed toward students in the first year of their baccalaureate (only), who have a special interest and talent in the fields related to the life sciences (primarily biology and chemistry).
Students may apply to a maximum of 3 programmes within the "Crazy About Science" series, and can participate in only one.
How to apply
Applications have to be submitted from here, starting on 12 September 2024.
Interested students must fill in the online application form and include a letter of motivation. A letter of recommendation will be requested directly from two of their teachers, who should know the applicant well. If the applicant has recently changed school then the letters of recommendation should be requested from his/her former teachers.
The deadline for registration is 18 October 2024 (23:59h).
The course is open to a total of 25 students. Candidates will be selected on the basis of their academic record, teacher recommendations and motivation to participate. A shortlist of candidates will be invited for interviews with the scientific organisers in November after which the final selection will be made. Students will be informed of the outcome by 25 November. The students selected to participate and their parents/guardians will be asked to sign a letter of commitment to attend all sessions.
In collaboration with
If you have any question, please contact us at irb_outreach@irbbarcelona.org
Important dates
- 18 October 2024: Registration deadline
- 4 November 2024: Contact with preselected candidates
- 18 November - 20 November 2024: Interviews
- 25 November 2024: Contact with selected candidates
- 10 January 2025: Official course inauguration
- 21 January 2025: Course start
- Closing ceremony - exact date to be confirmed
Programme
1. Destroy to discover the cure – Investigating Angelman syndrome
Karen Sfyaki (Targeted Protein Degradation and Drug Discovery Laboratory)

Angelman syndrome is a complex genetic disorder affecting primarily the brain. The main characteristics of this syndrome are delayed development, speech impairment, and problems with movement and balance and, in severe cases, epileptic seizures, and sleep disorders. Currently, there is no cure for Angelman syndrome, but we know that it is caused by the loss of a protein called UBE3A. This protein belongs to the family of E3 ubiquitin ligases. These enzymes mark proteins for degradation through the proteasome. The proteasome is the cell’s trash machinery that breaks down the proteins that are damaged or that are no longer needed. Therefore, without UBE3A, its substrates cannot be degraded by the proteasome and it will accumulate. However, the substrates of UBE3A are not yet known and they could be potential therapeutic targets for Angelman syndrome.
In recent years, a new application of the proteasome system has been developed to study proteins. Scientists have strategically managed to use the proteasome to their advantage by tricking it into degrading proteins that they are interested in. This approach is known as “targeted protein degradation”. To achieve this, we add a degradation tag to the protein of interest. This tag can be recognised by drugs called degraders, which bring the target protein and an E3 ligase closely together so that the protein can be marked for proteasomal degradation. This technology allows us to destroy any protein of interest and discover its downstream targets.
In this course, students will add the degradation tag to UBE3A using molecular cloning techniques and will become familiar with the techniques of DNA quantification, cloning and bacterial transformation.
2. Chemistry in a Click!: obtaining complex molecules through easy reactions
Ines Maria Garcia (Research Unit on Asymmetric Synthesis)

Chemists have always wanted to harness the power of molecular assembly to build complex molecules and obtain the widest range of applications. The logic behind this interest is simple—we are always in need of new molecular properties. These properties can be achieved by joining small molecular building blocks together—a process that becomes more difficult as the complexity of the molecules increases.
In 2001, Barry Sharpless introduced the concept of Click Chemistry as a new approach to synthesise complex drug-like molecules by using a few practical and reliable reactions. Click Chemistry includes chemical reactions that are performed with high selectivity, high yield, and a fast reaction rate, so that the “click” refers to the convenience afforded by snapping two objects together, like a luggage strap connection.
Click Chemistry has the following advantages over other types of reactions: it occurs in one pot; it is not disturbed by water; it generates minimal and usually harmless byproducts; and it has a high reaction specificity. Given these properties, this type of chemistry has been highly successful in recent years, leading its authors to win the Nobel Prize in Chemistry in 2022.
In this course, students will learn the basics of Click Chemistry and how to prepare these reactions. They will control these reactions using thin layer chromatography, detect the presence of both the reagents and the reaction products, and then finally purify the desired product.
3. Dealing with Stress – Learning survival mechanisms from the simplest eukaryotic organism
Ana Contreras & Ariadna Torres (Cell Signalling Laboratory)


A fundamental property of living cells is the ability to sense and respond to fluctuations in their environment. Cells have developed a large number of signal transduction pathways that serve to adapt to these changes. In our lab, we aim to unravel how cells detect, respond, and adapt to environmental variations.
Working with the budding yeast Saccharomyces cerevisiae, a well-established eukaryotic model organism that has the following advantages: ease of manipulation; short life span; ability to produce a large number of offspring; and a sequenced genome. We use S. cerevisiae to study the adaptive eukaryotic responses to a variety of environmental stresses. Remarkably, many processes initially discovered and studied in yeast, such as transcription regulation, stress-signalling transduction, and cell cycle regulation are conserved in higher eukaryotes.
Yeast and mammals share a conserved family of proteins known as Stress-Activated Protein Kinases (SAPKs) that sense and respond to environmental changes. The activation of SAPKs leads to a set of adaptive responses that involve the modulation of several physiological processes, including gene transcription, cell metabolism, protein translation, and cell cycle progression.
In this course, students will learn how to manipulate and work with S. cerevisiae to understand how cells detect and respond to external signals in order to ensure survival. For this purpose, we will study the activation of Hog1 SAPK upon different stress conditions and examine the cell fitness of various mutant strains when subjected to environmental insults.
4. Mirror, mirror on the wall… Which is the most helpful insect of them all? – Getting to understand tumorigenesis through the fly
Berta Muñoz, Carlos Escribano & Nikita Nikita (Development and Growth Control Laboratory)


The fruit fly Drosophila melanogaster has been widely used as a model organism for more than one hundred years to address biological questions in various fields. This organism has emerged as a potent tool for genetic manipulation, offering innumerable possibilities to analyse the detailed interaction between cells and tissues.
One question could be raised: how can a tiny organism like D. melanogaster be used to understand the pathways of a disease as complex as cancer? The answer is quite simple. Many biological mechanisms are well-conserved across evolution, including those related to pathological conditions. This conservation allows scientists to translate the knowledge acquired from less complex organisms to more complex ones. For instance, 75% of the genes related to human diseases and 68% of the genes related to human cancer have a counterpart in the fly. We are more similar to the fly
than we may think!
In this course, students will learn how to study the complex process of tumorigenesis using this model organism, both with a local and systemic approach. We will focus mainly on carcinomas, the most common type of tumour diagnosed in humans.
Carcinomas are derived from epithelial tissue, such as the skin, and they can become invasive or metastatic by spreading beyond the primary tissue layer and surrounding tissues or organs. In aggressive cancer cells, this transition is mediated by the activation of the EMT (Epithelial to Mesenchymal Transition) program, which causes the cells to undergo morphogenetic alterations that increase invasive capacity.
In this course, students will be introduced to the first steps in fly genetics. We will cross flies, identify genetic markers and balancer chromosomes, and apply other genetic tools we use in the lab every day to generate tumorigenic flies and study them. Students will also learn about the anatomy of the fly in adult and larvae stages and use advanced microscopy to detect several genetic markers in the tumoral
tissues. Finally, they will perform in vivo dissections and the respective immunohistochemistry.
5. Using the protein-eating machine to treat cancer
Jake Antony Ward (Targeted Protein Degradation and Drug Discovery Laboratory)

Control over the concentration, structure, and location of proteins is important for the health and fitness of our cells. When proteins are damaged or are no longer needed, they are broken down within the cell in a process known as degradation. One of the main cellular systems involved in degrading unwanted or damaged proteins is called the proteasome. The proteasome is made up of many enzymes, such as proteases, which recognise and break down these proteins. For a damaged or unwanted protein to be recognised and broken down by the proteasome, a specific label, known as ubiquitin, is usually added to the protein. Certain enzymes known as ubiquitin ligases are involved in this tagging process.
In recent years, drugs have been developed that use this proteasome system in our cells to degrade certain proteins. This is an exciting new strategy, as it can allow us to remove a given protein from a tumorogenic cell. This approach is known as Targeted Protein Degradation (TPD). This technology works by using a drug that brings the ubiquitin ligase and the target protein for degradation closer together. The protein is thus labelled with the ubiquitin-tag and is recognised by the proteasome and destroyed.
In this course, students will focus on a type of cancer known as Gastrointestinal Stromal Tumour (GIST). This cancer typically develops in the muscle cells of the gut. When mutated, the KIT protein is the molecule responsible for the formation and growth of this type of cancer. We are interested in designing drugs using the TPD strategy to remove KIT from GIST cancers. This line of research is exciting as it may lead to new treatments for GIST. Students will gain hands-on experience with the key techniques of gel electrophoresis and Western blotting to monitor the degradation of KIT in human cancer cells.
6. In vivo experimental approach to study breast cancer bone metastasis
Milos Lazic (Growth Control and Cancer Metastasis Laboratory) & Nerea González (Signalling and Cell Cycle Laboratory)


Metastasis is the process where cancer cells from a primary tumour try to enter the bloodstream, travel to other parts of the body, and form new tumours in different organs. Although many cancer cells attempt this, most
fail. However, sometimes these cells gain special abilities that allow them to spread and grow in new places.
Studying metastasis is difficult because it is a very complex process. We need a wide range of tools and techniques to understand it fully. Experiments in the lab (in vitro) help researchers learn about cancer cell behaviour in a controlled setting, but studying them in living organisms (in vivo) gives us a better understanding of how these cells spread and grow in the body.
In the laboratory, we use both methods to study how breast cancer spreads. We work with human cancer cells and animal models to investigate the role of specific genes in the metastasis process.
In this course, students will learn how to culture and label cells in the laboratory, monitor tumour growth, and analyse the results from in vivo experiments.
7. Machine learning to fight drug resistance
Gema Rojas & Elena Pareja (Structural Bioinformatics and Network Biology Laboratory)


Drug resistance, which occurs when previously vulnerable cells become unresponsive to specific drugs, poses a significant public health concern worldwide. Antimicrobial resistance (AMR) exemplifies this issue, with estimates indicating that the annual death toll resulting from AMR could skyrocket from 700,000 to 10 million people by 2050. Similarly, drug resistance affects the efficacy of cancer treatment, as patients may develop resistance following a relapse. The primary driver of drug resistance involves mutations in the DNA sequence of proteins associated with the drug's mode of action.
To comprehensively study this phenomenon, a crucial step involves analysing DNA sequencing data and establishing connections with protein structure and function to enhance our understanding of resistance mechanisms and devise strategies to overcome them. Machine learning has emerged as a valuable tool in predicting drug resistance by leveraging sequencing data.
In this course, students will have the opportunity to explore public databases and extract relevant biological information concerning drug resistance. They will work hands-on with sequencing data to understand the significance of mutations and observe how alterations in the sequence impact the shape and function of proteins. Finally, they will build a small machine-learning model to predict drug resistance on
the basis of sequence data.
8. CRISRP/Cas9 technology to generate a knockout in cancer cell lines
Camilla Bertani & Elsa Tusquets (Translational Control of Cell Cycle and Differentiation)

Our laboratory focuses on studying a family of proteins called Cytoplasmic Polyadenylation Element Binding Proteins (CPEBs), which are sequence-specific mRNA- binding proteins that control translation in development, health, and diseases such as cancer. A useful approach to evaluate the role of these proteins in cancer cells is to induce their deletion (knockout (KO)) and to evaluate the resulting phenotype.
In the laboratory, we have designed a genomic editing approach based on the CRISPR / Cas9 technique. We aim to induce the deletion of an exon present in the genomic sequence of these proteins in order to block their functionality. Before being able to evaluate the final phenotype, it is necessary to identify which clones obtained carry the exon deletion. Thus, in this course, students will analyse the DNA of these clones using a technique called PCR.
9. The light of evolution and the lab in a computer
Saioa Manzano (Comparative Genomics Laboratory)

All species on Earth are under constant change. Over generations, the DNA molecules of individuals within a population will undergo random changes (termed mutations), and this can sometimes translate to alterations in their biology, such as the emergence of a new trait (like resistance to a given antibiotic, or a colour that helps them camouflage better). If this trait is advantageous, individuals carrying the mutation will survive better and reproduce more than their counterparts, influencing the genetic makeup of the population. With time, this can result in a whole new species and it is the basis underlying the extraordinary biodiversity present on the planet.
DNA sequencing allows us to keep track of the emergence of these mutations across individuals and link them to the changes we can see in the clinic. By analyzing the similarities and differences in these sequences at a large scale (which we call comparative genomics), we can reconstruct the relationship between the populations or species carrying them, and understand how new traits appear and evolve. For example, this field of science proved very useful during the COVID-19 pandemic as it allowed researchers to monitor the emergence of new variants of SARS-CoV-2 and their spread across different countries almost in real-time, as well as develop vaccines that targeted proteins that were less prone to change.
In this course, students will apply a computational (dry lab) perspective to the understanding of the evolution of pathogenic agents. In particular, students will track the emergence of a new human virus from an animal pathogen and examine the changes that have allowed it to make the jump from one host to another, and we will gain an understanding of where it originated and how it spread. Students will learn how computers can be a powerful tool in a researcher's toolkit for understanding infectious diseases, predicting outbreaks, and designing therapeutic strategies.
10. Know your oral residents!
Mrinalini Parmar (Comparative Genomics Laboratory)

The healthy state of the human body is determined by numerous interacting factors. The central information comes from the cells’ genetic material, which serves as the primary determinant of its function and physiology. One aspect of this interaction involves the neighbouring cells, specifically microorganisms. The microbes in the human body are highly diverse and are found on most surfaces, including the gut, mouth, skin, and lungs. To illustrate their impact, there is approximately one microbe for every human cell, a fact only confirmed in 2016! For these microbes, the human body acts as an incubator, providing optimal survival conditions such as temperature and nutrients. However, little is known about how these neighbouring microbes influence human body function.
In recent years, this knowledge gap has put a spotlight on microbiome studies, which examine all the bacteria, fungi and viruses that reside in the human body. The microbiome field is relatively new, and its insights have shifted our understanding of health and disease. Recent studies have shown that microbes play key roles in digestion, immune system function, and even mental health. Current applications range from defining a healthy gut microbial composition for use in probiotics to treating irritable bowel syndrome (IBS) patients through faecal microbiota transplants.
In this course, students will get hands-on experience working with human samples of the oral microbiome, mainly the bacterial fauna. They will learn techniques ranging from isolation and identification of bacteria by colony morphology to confirmation of the species by amplicon sequencing of regions of 16S ribosomal RNA.
Venue
Crazy About Biomedicine course will take place at the IRB Barcelona facilities.
Institute for Research in Biomedicine (IRB Barcelona)
Parc Cientific de Barcelona
C/ Baldiri Reixac, 10
08028 Barcelona


