Organitzat per l'IRB Barcelona en col·laboració amb la Fundació Catalunya La Pedrera.
Presentation

Objectius
El Bojos per la Biomedicina és un curs dirigit als estudiants del primer any de batxillerat que desitgin explorar alguns dels descobriments fascinants que s'estan fent actualment en les ciències de la vida. A través d'aquest curs, els estudiants tindran l'oportunitat d'aprofundir el seu coneixement de la teoria i tècniques científiques en el camp de la biomedicina. Treballaran juntament amb investigadors joves per experimentar com es fa ciència en un institut de recerca internacional, guanyar una mica d'experiència pràctica en les últimes metodologies d'avantguarda i posicionar-se per a una possible carrera professional en les ciències de la vida.
Descripció del curs
Taller d'un any de durada sobre les ciències de la vida per a estudiants de batxillerat. Organitzat per l'IRB Barcelona dins el Programa "Bojos per la Ciència" de la Fundació Catalunya La Pedrera.
Aquest curs combina sessions teòriques i activitats experimentals pràctiques, que es duran a terme durant 16 dissabtes de l'any. El curs tractarà 10 temes científics actuals, que van des de la biologia cel·lular i molecular fins a la biologia estructural i computacional i la química, presentats per investigadors joves de l'IRB Barcelona. En el primer semestre (gener-abril), els tres primers dissabtes es dedicaran a aquestes sessions teòriques generals per a tots els participants. Durant els sis dissabtes següents, es formaran grups petits que entraran als laboratoris per a les sessions pràctiques. A continuació, es repetirà aquest programa amb 5 temes de recerca nous per al segon semestre (maig-novembre). Els estudiants participants s'hauran de comprometre a assistir al curs durant tot l'any.
Els estudiants rebran un certificat de participació en finalitzar el curs en una cerimònia especial de cloenda, a la que podran assistir pares i professors.
Idioma del curs
Totes les xerrades i sessions pràctiques es faran en anglès.

Dates i horaris
El curs tindrà lloc de gener a novembre de 2023, 10.00-14.00h.
1r SEMESTRE
- Diss. 21 gener: Benvinguda i xerrada 1
- Diss. 28 gener: Xerrades 2-3
- Diss. 18 febrer: Xerrades 4-5
- Diss. 4 març: Sessió pràctica 1
- Diss. 18 març: Sessió pràctica 2
- Diss. 1 abril: Sessió pràctica 3
- Diss. 22 abril: Sessió pràctica 4
- Diss. 13 maig: Sessió pràctica 5
2n SEMESTRE
- Diss. 20 maig: Xerrada 1
- Diss. 3 juny: Xerrades 2-3
- Diss. 10 juny: Xerrades 4-5
- Diss. 16 setembre: Sessió pràctica 1
- Diss. 30 setembre: Sessió pràctica 2
- Diss. 7 octubre: Sessió pràctica 3
- Diss. 21 octubre: Sessió pràctica 4
- Diss. 4 novembre: Sessió pràctica 5
Preu del curs
Els participants hauran d'abonar a la Fundació Catalunya La Pedrera la quantitat de 425 euros. Consulteu l'oferta de beques al seu lloc web.
Lloc on es realitzarà el curs
Institut de Recerca Biomèdica (IRB Barcelona)
C/ Baldiri Reixac, 10
08028 Barcelona
Qui pot sol·licitar una plaça
Aquest curs està dirigit als estudiants de primer any de batxillerat que tinguin un interès i talent especials en els camps relacionats amb les ciències de la vida.
Els estudiants poden sol·licitar plaça a un màxim de 3 dels programes de la sèrie "Bojos per la Ciència" i finalment només podran participar en un d'ells.
Com sol·licitar una plaça
Les inscripcions s'hauran de realitzar aquí a partir del 12 de setembre 2022
Els estudiants interessats hauran d'emplenar el formulari de sol·licitud i incloure una carta de motivació. També es demanarà una carta de recomanació directament de dos dels seus professors que coneguin bé l'alumne/a. En el cas de que l'estudiant hagi canviat de centre aquest curs, suggerim que sol·licitin les cartes als antics professors.
La data límit d'inscripció és el 19 d'octubre 2022 (23:59h).
El curs està obert a un total de 25 estudiants. Se seleccionaran els candidats en funció del seu expedient acadèmic, de les recomanacions dels seus professors i de la seva motivació per participar-hi. Es convidarà els candidats preseleccionats a fer entrevistes amb els organitzadors científics al novembre, després de les quals es farà la selecció final. La primera setmana de desembre es comunicarà el resultat als estudiants. Es demanarà als estudiants seleccionats per participar-hi i als seus pares/tutors legals que signin una carta de compromís d'assistir a totes les sessions.
Col·laboradors
Per a qualsevol dubte, si us plau contacteu-nos a irb_outreach@irbbarcelona.org
Dates importants
- 19 d'octubre 2022: Data límit inscripció
- 28 d’octubre 2022: Contacte amb els candidats pre-seleccionats
- 28 d’octubre -27 de novembre 2022: Entrevistes
- 28 de novembre 2022: Contacte amb els candidats seleccionats
- 13 de gener 2023: Inauguració oficial del curs
- 21 de gener 2023: Inici del curs
- Cerimònia de Clausura - data exacta a concretar
Programme
SEMESTER 1
1. 1. Unravelling Hematopoietic Stem Cells
Indranil Singh (Quantitative Stem Cells Dynamics)

Hematopoiesis depends on the preservation of a specific cell type called hematopoietic stem cells (HSCs). These long-lasting HSCs occupy the highest position in the hierarchy of hematopoietic cells and come with both self-renewal and pluripotent differentiation capacities. These HSCs differentiate into a full spectrum of mature blood cells through intermediate progenitor stages. While their long-term self-renewal potential helps in maintaining a source of primitive HSCs throughout one lifespan. At the same time, their ability to differentiate helps one to fight infection and disease. The balance between HSC self-renewal and differentiation is central to homeostasis and is regulated by the complex interplay of extrinsic and intrinsic regulatory networks.
As they age, HSCs gradually lose their differentiation potential. In our current effort, we are trying to understand the molecular mechanisms that contribute to HSC aging. We argue that most of the causes that underlie HSC aging result from cell-intrinsic pathways. Because many hematological pathologies are strongly age-associated, hence there has been an increasing interest in strategies to intervene in aspects of the functional stem cell expansion for transplantation or targeting the stem cell aging process could be of significant clinical relevance.
This course provides hands-on experience to learn the basic techniques to culture hematopoietic stem cells and they will be trained to induce ex-vivo differentiation. They will also have the opportunity to perform in vitro assays that will help you to recognize the cell types based on cell surface markers. During this workshop, you will be introduced to FACS (Flow activated cell sorting) concepts of suspension and adherent cell culture, media formation, and genomic DNA extractions.
2. Manipulating cellular plasticity
Isabel Calvo & Valentina Ramponi (Cellular Plasticity and Disease)


The concept of cellular plasticity has gained great relevance during the last years in the context of cancer and tissue repair. Cellular plasticity allows adult cells to regress to stem cell-like states through de-differentiation pathways.
It is possible to convert differentiated cells into pluripotent stem cells (induced pluripotent stem cells or iPSCs) by the simple expression of four transcription factors. These iPSCs are functionally equivalent to embryonic stem cells (ESCs), which are derived from the developing blastocyst and can divide indefinitely while maintaining the capacity to differentiate into any cell type of the organism.
There is an increasing interest in better understanding how these transitions occur both in vitro and in vivo and how they can be manipulated. This knowledge will be directly applied in regenerative medicine to improve current medical treatments.
Students will learn the basic techniques to culture differentiated and pluripotent stem cells and they will be trained to induce the transition between cellular states. Finally, they will have the opportunity to perform in vitro assays that will help us to recognize the ultimate state of pluripotency
3. CRISPR WARS: The Gene Strikes Back
Luis Povoas (Gene Translation Laboratory) 
Some years ago, during the process of constructing a fly model to study human diseases our lab found a very interesting protein that seems to be responsible for many different processes in the cell. This protein is a derivate of a group of proteins responsible to introduce amino acids in tRNA, to enable protein synthesis. We called it SLIMP (seryl-tRNA synthetase-like insect mitochondrial protein). Past studies suggest that this new protein has acquired an essential function in insects, being one of the most interesting results obtained so far, the role of SLIMP in cell cycle progression. To uncover more of SLIMP’s functions, our lab is using a revolutionary technology that changed the way we make science. This technology is the CRISPR editing system. This tool enables us to precisely edit the genome of live cells. With it, we can remove genes (knock-out), introduce genes (knock- in), introduce markers to follow a specific protein (tags) and others. Therefore, with this technology, we expect to uncover SLIMP intracellular location, protein-protein interactions and try to understand the SLIMP knockdown phenotypes in relation to DNA damage and cell cycle checkpoints. During this workshop, you will be introduced to concepts of molecular cloning, cell culture and CRISPR technology. May the science be with you!
4. From biomedicine to computational biology
Alba Sala (Molecular Modelling and Bioinformatics)

The interaction between small molecules is essential to living organisms. Amongst these molecules we find protein and nucleic acids, whose interaction plays a crucial role in biology (e.g., in the storage, repair, expression and regulation of genetic information).
Protein-DNA interaction is mostly defined by two mechanisms: what is called a direct or base read out and an indirect or shape readout. The former describes the ability of proteins to distinguish different binding modes. The later, focuses on the importance of certain protein and DNA sequences to adopt different 3D structures which determine the binding sites. The amount of importance given to these two mechanisms varies from protein to protein, but it is crucial to understand the role of a protein’s 3D structure.
In this course we will study the 3D structure of different proteins from a computational point of view. A protein’s structure is believed to dictate its function, and once a protein’s shape is understood, its role within the essential biological mechanisms can be determined; giving then information to scientists to develop new methodologies that work with a protein’s shape.
5. Introduction to Drosophila melanogaster research: a versatile model in biology & medicine
Bitarka Bisai (Development and Morphogenesis in Drosophila)
 The fruit fly, Drosophila melano gaster, is used as a model organism to study disciplines ranging from fundamental genetics to the development of tissues and organs. Despite the obvious differences between humans and Drosophila, it is remarkable that the fruit fly shares many similarities and conserved pathways with humans. Drosophila genome is 60% homologous to that of humans, less redundant, and about 75% of the genes responsible for human diseases have homologs in flies.
The fruit fly, Drosophila melano gaster, is used as a model organism to study disciplines ranging from fundamental genetics to the development of tissues and organs. Despite the obvious differences between humans and Drosophila, it is remarkable that the fruit fly shares many similarities and conserved pathways with humans. Drosophila genome is 60% homologous to that of humans, less redundant, and about 75% of the genes responsible for human diseases have homologs in flies.
Multipotency has been the focus of extensive work in biomedicine research. However, we lack much knowledge of multipotent cells in their physiological context in the full organism. During this workshop, we will address some of these issues by exploiting a naturally occurring population of multipotent cells, the adult progenitor cells (APCs) in Drosophila. In Drosophila, most larval cells, mainly polyploid, die at the transition between larval and adult stages; only the APCs survive and proceed into their terminal differentiation during metamorphosis. However, both APCs and larval cells are affected by the same nutritional and hormonal cues, suggesting that unique molecular components act in the adult progenitor cells to differentially regulate the effect of the external and the intrinsic stimuli in their unique setting.
This course provides hands-on experience of different techniques to understand Drosophila melanogaster research. You will be introduced to first steps in fly genetics through experimentation by crossing flies, identifying genetic markers and balancer chromosomes, and applying other genetic tools used every day in the lab. We will learn about the different stages of development and the anatomy of the fly at different stages. We will also perform fly dissections and use advanced microscopy.
SEMESTER 2
1. Un Dealing with Stress – Learning survival mechanisms from the simplest eukaryotic organism
Iván Company, Maria Quintana, Guillem Pérez & Ariadna Torres (Cell Signalling)



A fundamental property of living cells is the ability to sense and respond to fluctuations in their environment. Cells have developed a wide number of signal transduction pathways that serve to adapt to these changes. In our lab, we aim to unravel how cells detect, respond and adapt to environmental variations.
The budding yeast Saccharomyces cerevisiae, a well-established eukaryotic model organism, has several advantages: ease of manipulation, short life span, ability to produce a large number of offspring and a sequenced genome. We use S. cerevisiae to study the adaptive eukaryotic responses to a variety of environmental stresses. Remarkably, many processes initially discovered and studied in yeast, are conserved in higher eukaryotes, such as transcription regulation, stress-signaling transduction and cell cycle regulation.
Yeast and mammals share a conserved family of proteins that sense and respond to environmental changes known as Stress-Activated Protein Kinases (SAPKs). The activation of SAPKs leads to a set of adaptive responses that involve the modulation of several physiological processes like changes in gene transcription, cell metabolism, protein translation and cell cycle progression.
In this practical course, we will learn how to manipulate and work with S. cerevisiae to understand how cells detect and respond to external signals in order to ensure survival. For this purpose, we will study at the protein level the activation of Hog1 SAPK upon different stress conditions and examine the cell fitness of different mutant strains when subjected to environmental insults.
2. Mirror, Mirror on the Wall… Who Is the Most Helpful Insect of Them All? – Getting to Know Tumorigenesis Through the Fly
Elena Fusari & Amanda González (Development and Growth Control Laboratory)
The fruit fly Drosophila melanogaster has been widely used as a model organism for more than one hundred years to address biological questions in various fields. This organism has emerged as a potent tool for genetic manipulation, offering innumerable possibilities to analyze the detailed interaction between cells and tissues.
One question could be raised: how can a tiny organism such as Drosophila melanogaster be used to understand the pathways of such a complex disease as cancer? Well, the answer is as simple as that: many biological mechanisms are well conserved across evolution, including the ones concerning pathological conditions. This is what allows scientists to translate the knowledge acquired from less complex organisms to more complex ones. For instance, 75% of human disease-related genes and 68% of human cancer-related genes have a counterpart in the fly. We are more similar to the fly than what we may think!
During this semester, we will learn how to study the complex process of tumorigenesis using this model organism, both with a local and systemic approach. We will focus mainly on carcinomas, the most common type of tumor diagnosed in humans.
Carcinomas are derived from epithelial tissue, such as the skin, and they can become invasive or metastatic by spreading beyond the primary tissue layer and surrounding tissues or organs. In aggressive cancer cells, this transition is mediated by the activation of the EMT (Epithelial to Mesenchymal Transition) program, which causes the cells to undergo morphogenetic alterations that increase invasive capacity.
In this course, we will introduce you to the first steps in fly genetics. We will cross flies, identify genetic markers and balancer chromosomes, and apply other genetic tools that we use in the lab every day in order to generate tumorigenic flies and study them. We will also learn about the anatomy of the fly in adult and larvae stages and use advanced microscopy to see several genetic markers in the tumoral tissues. We will also perform in vivo dissections and their respective immunohistochemistry.
3. NMR to study molecules in solution
Miriam Condeminas (Structural Characterization of Macromolecular Assemblies)
Structural Biology aims at understanding the shape and function of biological molecules to decipher the basic mechanisms of life. It can also help identify molecules with pharmacological applications to treat human diseases. Nowadays, such questions can be answered using a wide variety of complementary tools that include Nuclear Magnetic Resonance (NMR).
In NMR, samples are irradiated with radio waves while placed inside a strong magnetic field. As a result, frequency changes in the nuclei of the molecules can be recorded and the corresponding signals plotted into NMR spectra. By combining various types of NMR experiments, each signal can be linked to a particular nucleus and thus yield important information regarding its chemical environment. Given that these experiments can be performed in aqueous solutions, NMR is very versatile in that it enables the observation of macromolecular flexible regions, as well as dynamic processes (invisible using other techniques) and it can also map binding interfaces.
In this course, students will learn the fundamentals behind NMR, how to interpret the spectra resulting from various experiments and the workflow required for assigning a peptide.
4. An omics complex puzzle? Let’s decipher it by using Bioinformatics!
Olfat Khannous (Comparative Genomics) & Marina Murillo (Gene Translation Laboratory)

 Complex biological diseases such as cancer and Alzheimer are caused by a combination of genetic and environmental factors, and in many cases, we have not yet identified or established a relationship between them to shed light to the understanding of the origin and progression of such biological problems.
Complex biological diseases such as cancer and Alzheimer are caused by a combination of genetic and environmental factors, and in many cases, we have not yet identified or established a relationship between them to shed light to the understanding of the origin and progression of such biological problems.
Multi-omics profiling and integration combines different types of data for a better understanding of these complex diseases.
One of the branches of the ‘Omics’ sciences is transcriptomics which involves the study of all the RNA molecules (including mRNA, tRNA, rRNA, and other non-coding RNAs) within a cell, otherwise known as the transcriptome. Consequently, by analyzing the transcriptome researchers can determine if a gene is turned on or off in the cells and tissues of an organism.
On the other hand, metagenomics is the study of the genetic material (genomes) from a mixed community of organisms of a sample. It allows us to describe which microbial communities are living within each individual (microbiome) and its relationship with host genetics, diet and environment can reveal links with disease or health.
In our workshop you will be immersed in the computational world (dry lab) understanding its synergy with the experimental lab (wet lab), and you will learn how to obtain transcriptomics and metagenomics data and also how to use bioinformatics tools to analyze them and to add missing pieces to these mentioned complex puzzles.
5. CRISPR/Cas9 to generate a KO
Camilla Bertani (Translational Control of Cell Cycle and Differentiation)

Our laboratory is focused on studying a family of proteins called Cytoplasmic Polyadenylation Element Binding Proteins (CPEBs), which are sequence-specific mRNA- binding proteins that control translation in development, health and diseases, such as cancer. A useful approach to evaluate the role of these proteins in cancer cells is to induce their deletion (knockout (KO)) and to evaluate the resulting phenotype.
For this reason, in the laboratory we have designed a possible genomic editing approach, based on the CRISPR / Cas9 technique. Our aim is to induce the deletion of an exon present in the genomic sequence of these proteins in order to block their functionality. Before being able to evaluate the final phenotype, it is necessary to evaluate which clones obtained presents the deletion of the exon. Thus, in this workshop, we will analyze the DNA of these clones using a technique called PCR.
Venue
El curs de Bojos per la Biomedicina tindrà lloc a les instal·lacions de l'IRB Barcelona.
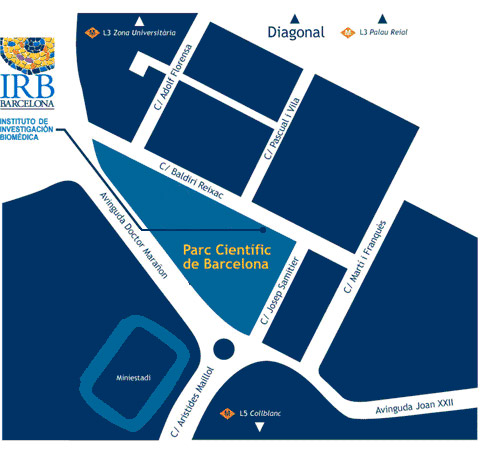
Institute for Research in Biomedicine (IRB Barcelona)
Parc Cientific de Barcelona
C/ Baldiri Reixac, 10
08028 Barcelona


