Organized by IRB Barcelona in collaboration with the Fundació Catalunya La Pedrera.
Presentation

Objectives
Crazy About Biomedicine is a course directed at students in their first year of baccalaureate who wish to explore some of the exciting discoveries being made in the life sciences. Through this course, students will have a chance to deepen their knowledge of scientific theory and techniques in the field of biomedicine. They will work alongside our young researchers to get a taste for what doing science in a top international research institute is like, gain some hands-on experience in the latest cutting-edge methodologies, and position themselves for a potential career in the life sciences.
Course Description
A year-long workshop in the life sciences for high-school students organized by IRB Barcelona within the series "Crazy About Science" of the Fundació Catalunya La Pedrera.
This course includes a combination of theoretical lectures and practical hands-on experimental activities, which take place on 16 Saturdays throughout the year. Presented by IRB Barcelona PhD students and postdocs, the course will cover 10 hot scientific topics, ranging from cell and molecular biology to structural and computational biology, and chemistry. In the first ‘semester’ (January-April), the first three Saturdays will be devoted to these general lectures for all participants. During the following five sessions, small groups will enter the labs for hands-on practical experience. This schedule will then be repeated with five new research topics in the second semester (May-November). Participants must commit themselves to attend the whole course.
Students will receive a Certificate of Participation upon successful completion of the course at a special ceremony to which parents and teachers will be invited.
Course language
All lectures and practical sessions will be conducted in English.

Course dates and times
The course will run from January to November 2022, Saturdays 10.00-14.00h.
SEMESTER I
- Fri. 14 January 2022: Official course inauguration
- Sat. 22 January: Welcome and talk 1
- Sat. 5 February: Talk 2-3
- Sat. 19 February: Talk 4-5
- Sat. 26 February: Practical session 1
- Sat. 12 March: Practical session 2
- Sat. 26 March: Practical session 3
- Sat. 2 April: Practical session 4
- Sat. 30 April: Practical session 5
SEMESTER II
- Sat. 21 May: Talk 1
- Sat. 4 June: Talk 2-3
- Sat. 11 June: Talk 4-5
- Sat. 17 September: Practical session 1
- Sat. 1 October: Practical session 2
- Sat. 15 October: Practical session 3
- Sat. 22 October: Practical session 4
- Sat. 5 November: Practical session 5
Course fees
There is a fee of 425 euros payable directly to the Fundació Catalunya La Pedrera. Take a look at the scholarships available at their webpage.
Course location
Institute for Research in Biomedicine (IRB Barcelona)
C/ Baldiri Reixac, 10
08028 Barcelona
Who can apply
This course is directed toward students in the first year of their baccalaureate (only), who have a special interest and talent in the fields related to the life sciences (primarily biology and chemistry).
Students may apply to a maximum of 3 programmes within the "Crazy About Science" series, and can participate in only one.
How to apply
Applications have to be submitted from here, starting on 15 September 2021.
Interested students must fill in the online application form and include a letter of motivation. A letter of recommendation will be requested directly from two of their teachers, who should know the applicant well. If the applicant has recently changed school then the letters of recommendation should be requested from his/her former teachers.
The deadline for registration is 25 October 2021 (23:59h).
The course is open to a total of 25 students. Candidates will be selected on the basis of their academic record, teacher recommendations and motivation to participate. A shortlist of candidates will be invited for interviews with the scientific organisers in November after which the final selection will be made. Students will be informed of the outcome by the first week in December. The students selected to participate and their parents/guardians will be asked to sign a letter of commitment to attend all sessions.
In collaboration with
Facebook: @LaPedrera.Fundacio
Twitter: @PedreraFundacio
Instagram: @lapedrera_fundacio
Facebook: @LaPedrera.Ciencia
Twitter: @PedreraScience
Instagram: @lapedrera_scienceacademy

If you have any question, please contact us at irb_outreach@irbbarcelona.org
Important dates
- 25 October 2021: Application deadline
- 12 November 2021: Short-listed candidates contacted
- 13-28 November 2021: Interviews
- 30 November 2021: Selected students contacted
- 14 January 2022: Official inauguration of the course
- 22 January 2022: Course begins
- 15 December 2022: Closing Ceremony
Programme
SEMESTER 1
1. Unravelling the Molecular Structure of Life
Blazej Baginski (Structural Characterization of Macromolecular Assemblies)

Proteins and nucleic acids (DNA and RNA) are the basic building blocks of life. They perform a multitude of functions–from sensing, transporting, and enzymatic regulation, to building the cell’s internal skeleton. Therefore, the fold and 3D-structure of these biomolecules is carefully controlled. A protein’s 3D structure determines its activity, creates receptor binding pockets and enzyme active centres.
Crystallography is one of the few methods that allows the structural determination of such macromolecules with atomic precision. By studying the interactions of crystallised molecules by means of high energy X-rays, it is possible to pinpoint the location of atoms and their bonds in a given molecule of interest.
During this course, we will set up a protein crystallisation experiment, learn the operation of high-precision pipetting robots, and cryogenically freeze protein crystals to prepare them for X-ray data collection at the synchrotron.
2. Manipulating cellular plasticity
Isabel Calvo (Cellular Plasticity and Disease)

The concept of cellular plasticity has gained great relevance during the last years in the context of cancer and tissue repair. Cellular plasticity allows adult cells to regress to stem cell-like states through de-differentiation pathways.
It is possible to convert differentiated cells into pluripotent stem cells (induced pluripotent stem cells or iPSCs) by the simple expression of four transcription factors. These iPSCs are functionally equivalent to embryonic stem cells (ESCs), which are derived from the developing blastocyst and can divide indefinitely while maintaining the capacity to differentiate into any cell type of the organism.
There is an increasing interest in better understanding how these transitions occur both in vitro and in vivo and how they can be manipulated. This knowledge will be directly applied in regenerative medicine to improve current medical treatments.
Students will learn the basic techniques to culture differentiated and pluripotent stem cells and they will be trained to induce the transition between cellular states. Finally, they will have the opportunity to perform in vitro assays that will help us to recognize the ultimate state of pluripotency.
3. CRISPR WARS: The Gene Strikes Back
Luis Povoas (Gene Translation Laboratory) 
Some years ago, during the process of constructing a fly model to study human diseases our lab found a very interesting protein that seems to be responsible for many different processes in the cell. This protein is a derivate of a group of proteins responsible to introduce amino acids in tRNA, to enable protein synthesis. We called it SLIMP (seryl-tRNA synthetase-like insect mitochondrial protein). Past studies suggest that this new protein has acquired an essential function in insects, being one of the most interesting results obtained so far, the role of SLIMP in cell cycle progression. To uncover more of SLIMP’s functions, our lab is using a revolutionary technology that changed the way we make science. This technology is the CRISPR editing system. This tool enables us to precisely edit the genome of live cells. With it, we can remove genes (knock-out), introduce genes (knock-in), introduce markers to follow a specific protein (tags) and others. Therefore, with this technology, we expect to uncover SLIMP intracellular location, protein-protein interactions and try to understand the SLIMP knockdown phenotypes in relation to DNA damage and cell cycle checkpoints. During this workshop, you will be introduced to concepts of molecular cloning, cell culture and CRISPR technology. May the science be with you!
4. From biomedicine to computational biology
Alba Sala (Molecular Modelling and Bioinformatics)

The interaction between small molecules is essential to living organisms. Amongst these molecules we find protein and nucleic acids, whose interaction plays a crucial role in biology (e.g., in the storage, repair, expression and regulation of genetic information).
Protein-DNA interaction is mostly defined by two mechanisms: what is called a direct or base read out and an indirect or shape readout. The former describes the ability of proteins to distinguish different binding modes. The later, focuses on the importance of certain protein and DNA sequences to adopt different 3D structures which determine the binding sites. The amount of importance given to these two mechanisms varies from protein to protein, but it is crucial to understand the role of a protein’s 3D structure.
In this course we will study the 3D structure of different proteins from a computational point of view. A protein’s structure is believed to dictate its function, and once a protein’s shape is understood, its role within the essential biological mechanisms can be determined; giving then information to scientists to develop new methodologies that work with a protein’s shape.
5. Introduction to Drosophila melanogaster research: a versatile model in biology & medicine
Bitarka Bisai (Development and Morphogenesis in Drosophila)
 The fruit fly, Drosophila melanogaster, is used as a model organism to study disciplines ranging from fundamental genetics to the development of tissues and organs. Despite the obvious differences between humans and Drosophila, it is remarkable that the fruit fly shares many similarities and conserved pathways with humans. Drosophila genome is 60% homologous to that of humans, less redundant, and about 75% of the genes responsible for human diseases have homologs in flies.
The fruit fly, Drosophila melanogaster, is used as a model organism to study disciplines ranging from fundamental genetics to the development of tissues and organs. Despite the obvious differences between humans and Drosophila, it is remarkable that the fruit fly shares many similarities and conserved pathways with humans. Drosophila genome is 60% homologous to that of humans, less redundant, and about 75% of the genes responsible for human diseases have homologs in flies.
Multipotency has been the focus of extensive work in biomedicine research. However, we lack much knowledge of multipotent cells in their physiological context in the full organism. During this workshop, we will address some of these issues by exploiting a naturally occurring population of multipotent cells, the adult progenitor cells (APCs) in Drosophila. In Drosophila, most larval cells, mainly polyploid, die at the transition between larval and adult stages; only the APCs survive and proceed into their terminal differentiation during metamorphosis. However, both APCs and larval cells are affected by the same nutritional and hormonal cues, suggesting that unique molecular components act in the adult progenitor cells to differentially regulate the effect of the external and the intrinsic stimuli in their unique setting.
This course provides hands-on experience of different techniques to understand Drosophila melanogaster research. You will be introduced to first steps in fly genetics through
experimentation by crossing flies, identifying genetic markers and balancer chromosomes, and applying other genetic tools used every day in the lab. We will learn about the different stages of development and the anatomy of the fly at different stages. We will also perform fly dissections and use advanced microscopy.
SEMESTER 2
1. Strategies to Understand Aging and Senescence
Marta Kovatcheva & Valentina Ramponi (Cellular Plasticity and Disease)


Scientific research and modern medicine have dramatically extended life expectancy, with the average person in the developed world expected to reach 80 years of age or more. However, this extension in lifespan has had no effect on health span; that is, the number of healthy years a human lives. Aging is still characterised by multiple pathologies including frailty, heart disease, cancer, and neurodegenerative diseases, among many others.
One of the main hallmarks of ageing is cellular senescence, the phenomenon by which normal cells stop dividing. Senescent cells accumulate in an organism over time, secreting pro-inflammatory molecules and contributing to age-related diseases. There is an increasing interest in clinical medicine to better identify and target senescent cells, as their elimination may delay and ameliorate some age-associated diseases.
This course provides hands-on experience using different techniques to induce cellular senescence in normal cells. We will learn state-of-the-art techniques to study the molecular biology of senescent cells, analysing in vitro and in vivo samples. Finally, we will perform classical protocols, such as SAβgal staining, to detect senescent cells. These techniques will help us to understand, identify and target senescent cells for clearance, which is a promising therapeutic approach to extend health span.
2. Mirror, Mirror on the Wall… Who Is the Most Helpful Insect of Them All? – Getting to Know Tumorigenesis Through the Fly
Elena Fusari & Amanda González (Development and Growth Control Laboratory)
The fruit fly Drosophila melanogaster has been widely used as a model organism for more than one hundred years to address biological questions in various fields. This organism has emerged as a potent tool for genetic manipulation, offering innumerable possibilities to analyze the detailed interaction between cells and tissues.
One question could be raised: how can a tiny organism such as Drosophila melanogaster be used to understand the pathways of such a complex disease as cancer? Well, the answer is as simple as that: many biological mechanisms are well conserved across evolution, including the ones concerning pathological conditions. This is what allows scientists to translate the knowledge acquired from less complex organisms to more complex ones. For instance, 75% of human disease-related genes and 68% of human cancer-related genes have a counterpart in the fly. We are more similar to the fly than what we may think!
During this semester, we will learn how to study the complex process of tumorigenesis using this model organism, both with a local and systemic approach. We will focus mainly on carcinomas, the most common type of tumor diagnosed in humans.
Carcinomas are derived from epithelial tissue, such as the skin, and they can become invasive or metastatic by spreading beyond the primary tissue layer and surrounding tissues or organs. In aggressive cancer cells, this transition is mediated by the activation of the EMT (Epithelial to Mesenchymal Transition) program, which causes the cells to undergo morphogenetic alterations that increase invasive capacity.
In this course, we will introduce you to the first steps in fly genetics. We will cross flies, identify genetic markers and balancer chromosomes, and apply other genetic tools that we use in the lab every day in order to generate tumorigenic flies and study them. We will also learn about the anatomy of the fly in adult and larvae stages and use advanced microscopy to see several genetic markers in the tumoral tissues. We will also perform in vivo dissections and their respective immunohistochemistry.
3. NMR to study molecules in solution
Miriam Condeminas (Structural Characterization of Macromolecular Assemblies)
Structural Biology aims at understanding the shape and function of biological molecules to decipher the basic mechanisms of life. It can also help identify molecules with pharmacological applications to treat human diseases. Nowadays, such questions can be answered using a wide variety of complementary tools that include Nuclear Magnetic Resonance (NMR).
In NMR, samples are irradiated with radio waves while placed inside a strong magnetic field. As a result, frequency changes in the nuclei of the molecules can be recorded and the corresponding signals plotted into NMR spectra. By combining various types of NMR experiments, each signal can be linked to a particular nucleus and thus yield important information regarding its chemical environment. Given that these experiments can be performed in aqueous solutions, NMR is very versatile in that it enables the observation of macromolecular flexible regions, as well as dynamic processes (invisible using other techniques) and it can also map binding interfaces.
In this course, students will learn the fundamentals behind NMR, how to interpret the spectra resulting from various experiments and the workflow required for assigning a peptide.
4. An omics complex puzzle? Let’s decipher it by using Bioinformatics!
Olfat Khannous (Molecular Modelling and Bioinformatics) & Marina Murillo (Gene Translation Laboratory)

 Complex biological diseases such as cancer and Alzheimer are caused by a combination of genetic and environmental factors, and in many cases we have not yet identified or established a relationship between them to shed light to the understanding of the origin and progression of such biological problems.
Complex biological diseases such as cancer and Alzheimer are caused by a combination of genetic and environmental factors, and in many cases we have not yet identified or established a relationship between them to shed light to the understanding of the origin and progression of such biological problems.
Multi-omics profiling and integration combines different types of data for a better understanding of these complex diseases.
One of the branches of the ‘Omics’ sciences is transcriptomics which involves the study of all the RNA molecules (including mRNA, tRNA, rRNA, and other non-coding RNAs) within a cell, otherwise known as the transcriptome. Consequently, by analyzing the transcriptome researchers can determine if a gene is turned on or off in the cells and tissues of an organism.
On the other hand, metagenomics is the study of the genetic material (genomes) from a mixed community of organisms of a sample. It allows us to describe which microbial communities are living within each individual (microbiome) and its relationship with host genetics, diet and environment can reveal links with disease or health.
In our workshop you will be immersed in the computational world (dry lab) understanding its synergy with the experimental lab (wet lab), and you will learn how to obtain transcriptomics and metagenomics data and also how to use bioinformatics tools to analyze them and to add missing pieces to these mentioned complex puzzles.
5. CRISPR/Cas9 to generate a KO
Camilla Bertani (Translational Control of Cell Cycle and Differentiation)

My laboratory is focused on studying a family of proteins called Cytoplasmic Polyadenylation Element Binding Proteins (CPEBs), are sequence-specific mRNA-binding proteins that control translation in development, health and diseases such as cancers. To evaluate the role these proteins, play in cancer cells, a good approach is to induce KO of the protein and evaluate the resulting phenotype.
For this reason, in the laboratory we have designed a possible genomic editing approach, based on the CRIPR / Cas9 technique. Our aim is to induce the deletion of an exon present in the genomic sequence of these proteins, in order to block their functionality.
Therefore, before being able to evaluate the phenotype, it is necessary to evaluate which of the clones we have obtained really presents the deletion of the exon and also, to evaluate if in homozygous or heterozygous form.
For this reason, it is essential to conduct the screening of clones, in particular the analysis of genomic DNA, through the use of a technique called PCR.
.
Venue
Crazy About Biomedicine course will take place at the IRB Barcelona facilities.
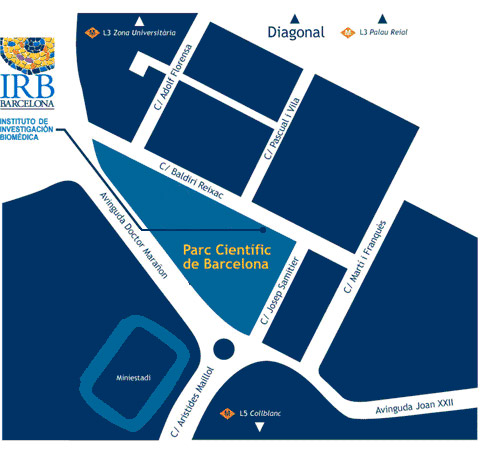
Institute for Research in Biomedicine (IRB Barcelona)
Parc Cientific de Barcelona
C/ Baldiri Reixac, 10
08028 Barcelona

